Characterization of ligand signaling signatures and biased agonism with bioSensAll®
Classically, GPCRs have been considered as binary signaling switches existing in equilibrium between “on” and “off” states. In this view, ligands are classified as agonists, inverse agonists or antagonists depending on their overall effect on this equilibrium (i.e., agonists shifting the equilibrium to the on state
and inverse agonists shifting the equilibrium to the off state). Two key complementary discoveries in the last decade, namely pluridimensionality of signaling efficacy and biased agonism [1], have revolutionized our understanding of GPCR biology and the pharmacology of ligands targeting these receptors. Contrary to the classical view of GPCR signaling, it is now evident that these receptors can exists in multiple active (i.e., “on”) states [2] and engage multiple (potentially crosstalking) downstream signaling cascades by directly coupling to both heterotrimeric G proteins and non-G protein effectors (i.e., β-arrestins). Further, a given drug acting on a receptor can display different (sometimes reversed) efficacies towards the various pathways coupled to the receptor [3]. is described by the concept of biased. Drug efficacy is thus considered to be a pluridimensional parameter.
An extension of this observation agonism (or functional selectivity), which posits that structurally dissimilar ligands can stabilize unique receptor conformations with each conformation differentially coupling to downstream effectors. Accordingly, different ligands of a given receptor may impart distinct signaling signatures and biological properties to the receptor. The promise of biased agonism lies in its applicability for the development of GPCR ligands that selectively engage therapeutic pathways while inhibiting or remaining inert towards those producing deleterious outcomes. Ultimately,
biased ligands may display superior therapeutic profiles in many disease areas and indications [4].
In this application note, we demonstrate how bioSensAll® biosensors were used to characterize and differentiate the signaling signatures of endogenous and synthetic human parathyroid hormone 1 receptor (hPTH1R) ligands (i.e., parathyroid hormone (PTH) and Tyr1 -PTH, respectively).
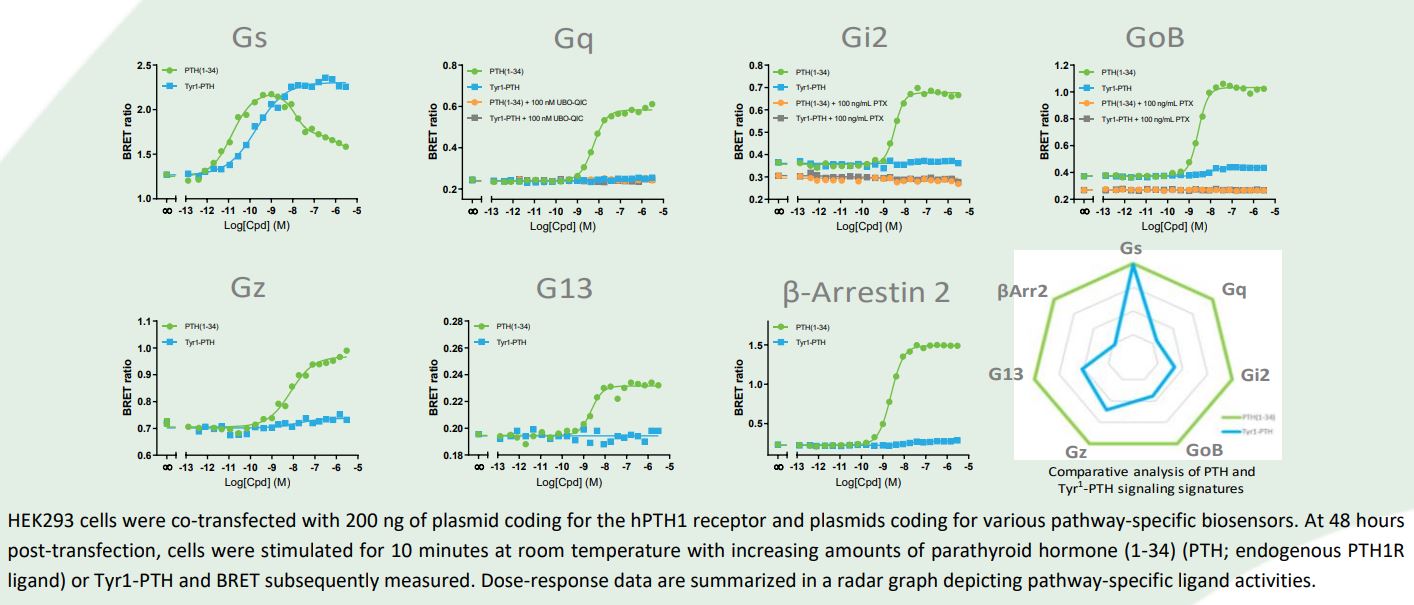
Results and Conclusion
In response to its endogenous ligand PTH, hPTH1R engaged Gɑs, Gɑq, Gi-family G-proteins (i.e.,
Gɑi2, GɑoB, Gɑz), Gɑ13 and β-arrestin 2. Interestingly, the synthetic ligand Tyr1-PTH only activated Gɑs (full agonist) while remaining relatively ineffective at stimulating the other pathways engaged by PTH. The bioSensAll® biosensor platform provides a qualitative and quantitative assessment of a receptor’s complete signaling repertoire and allows for the identification and characterization of biased ligands in a rapid, simple and homogenous assay. Ultimately, a ligand’s signaling signature may be useful in forecasting its therapeutic efficacy and safety.
References
[1] Kenakin, T. (2011) Functional selectivity and biased receptor signaling. J Pharmacol Exp Ther. 336(2):296-302.
[2] Ye, L., Van Eps, N., Zimmer, M., Ernst, O.P., Scott Prosser, R. (2016) Activation of the A2A adenosine
G-protein-coupled receptor by conformational selection. Nature. 533:265-68.
[3] Galandrin, S., Bouvier, M. (2006) Distinct signaling profiles of beta1 and beta2 adrenergic receptor ligands toward adenylyl cyclase and mitogen-activated protein kinase reveals the pluridimensionality of efficacy. Mol Pharmacol. 70(5):1575-84. [4] Kenakin, T., Christopoulos, A. (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 12(3):205-16.