Ligand profiling in cell lines of different tissular origin using bioSensAll®
Signaling via G protein-coupled receptors (GPCRs) is a complex multistep process that begins with a ligand-induced conformational rearrangement of the receptor and subsequent activation of receptor-coupled heterotrimeric G proteins and/or β-arrestins. It is now well-appreciated that different ligands of a
given GPCR can stabilize distinct receptor conformations, each provoking unique signaling profiles. The biological outcome of receptor activation is ultimately influenced by the complement of signaling and regulatory proteins present in a given cell type. This GPCR-related signaling “machinery” can vary in a cell typespecific manner [2] and ultimately influence the pharmacological properties (i.e., potency, efficacy) of a ligand. Such “system (or cell) bias” is especially prominent when using downstream second messenger levels as readouts, the latter being particularly sensitive to a cell’s signaling protein
repertoire. Importantly, cell bias complicates the identification of biased ligands and receptors.
In this application note, we demonstrate how proximal bioSensAll® assays can be used for profiling various CB1 receptor ligands in four mammalian cell lines commonly used in basic GPCR research: HEK-293, CHO-K1, HeLa and U2OS.
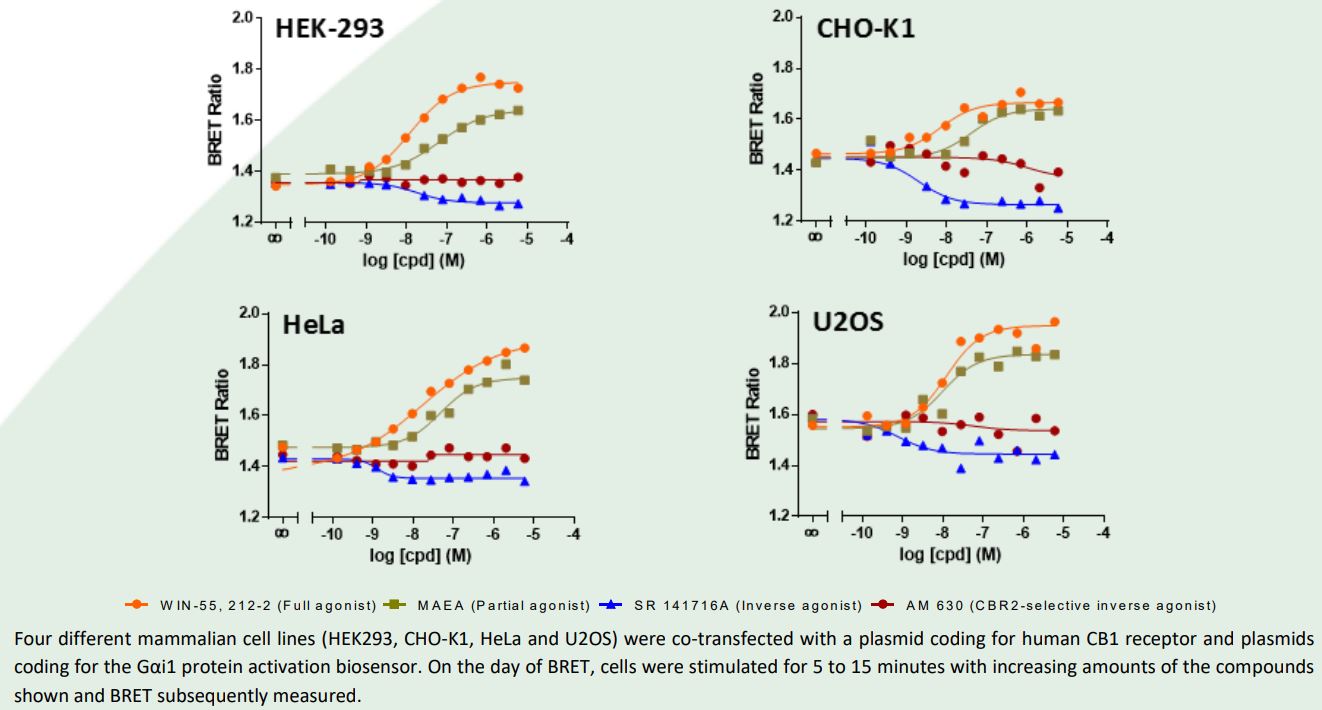
Results and Conclusion
CB1 receptor agonist potencies and relative efficacies were conserved among the four mammalian
cell lines tested. Further, compound SR 141716A retained its inverse agonist activity in all cell systems, while the CB2 receptor-specific inverse agonist AM-630 was devoid of activity in the four cell lines. Overall, these data reveal that CB1 receptor ligand pharmacology is well conserved across four distinct mammalian cell lines when measured using bioSensAll® G protein activation biosensors. The use of receptor-proximal measures of GPCR activity (i.e., G protein activation) can thus possibly render ligand pharmacology studies refractive to system/cell bias. Such bias may complicate ligand pharmacology studies performed with more traditional second messenger assays and may prove misleading in identifying ligand- and receptor-inherent bias.
bioSensAll® is a versatile technology that can be adapted to numerous cellular models for the study of receptor signaling and ligand pharmacology. Furthermore, by providing upstream measures of ligand activity, bioSensAll® decomplexifies the task of identifying ligand/receptor bias.
References
[1] Mancini, A., Frauli, M., Breton, B. (2015) Exploring the Technology Landscape of 7TMR Drug Signaling Profiling. Curr. Top. Med. Chem. 15(24):2528-42.
[2] Atwood, B.K., Lopez, J., Wager-Miller, J., Mackie, K., Straiker, A. (2011) Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis.
BMC Genomics. 12(14).
[3] Steen, A., Larsen, O., Thiele, S., Rosenkilde, M.M. (2014) Biased and G protein-independent signaling of chemokine receptors. Front Immunol. 23(5):277.