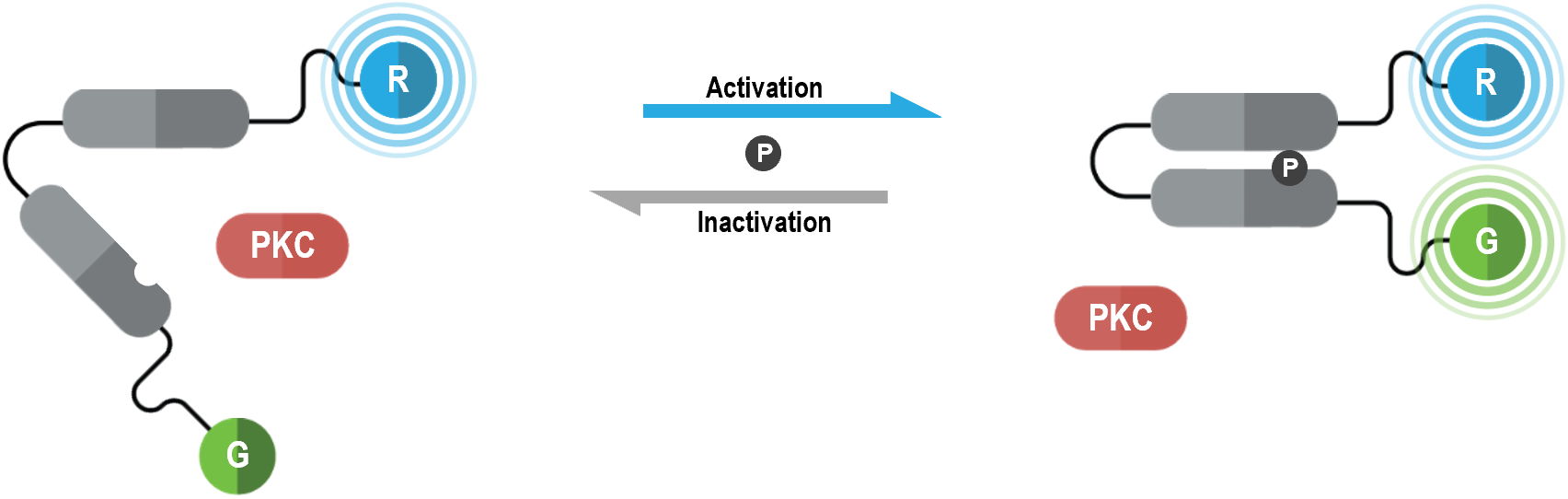
Distal effector monitoring: The PKC biosensor
How the biosensor works
The principal effector of the Gαq/11 pathway is phospholipase C-β (PLCβ), which catalyzes the cleavage of membrane-bound phosphatidylinositol 4,5-biphosphate (PIP2) into the second messengers inositol (1,4,5) trisphosphate (IP3) and diacylglycerol (DAG). Binding of IP3 to its receptor on the endoplasmic reticulum (ER) results in the release of ER calcium stores. Increased concentrations of calcium and DAG lead to the activation of protein kinase C (PKC). Consequently, biosensors monitoring DAG levels and PKC activity have been developed as distal measures of G protein-coupled receptor (GPCR)-downstream Gαq/11 activation.
The bioSens-All® unimolecular PKC activation sensor consists of “core region” containing PKC substrate domains fused via a flexible linker. This core region is tagged with Renilla luciferase (RLuc; R in figure below) at its N-terminus and green fluorescent protein (GFP; G in following figure) at its C-terminus. Upon PKC phosphorylation of the core region, the PKC substrate domains interact leading to increased physical proximity between N- and C-terminal RLuc and GFP tags. As a result, PKC activation translates into an increased BRET efficiency, the magnitude of the BRET signal being directly proportional to the level of PKC activity.
PKC biosensor data generated with the Bradykinin B2 receptor
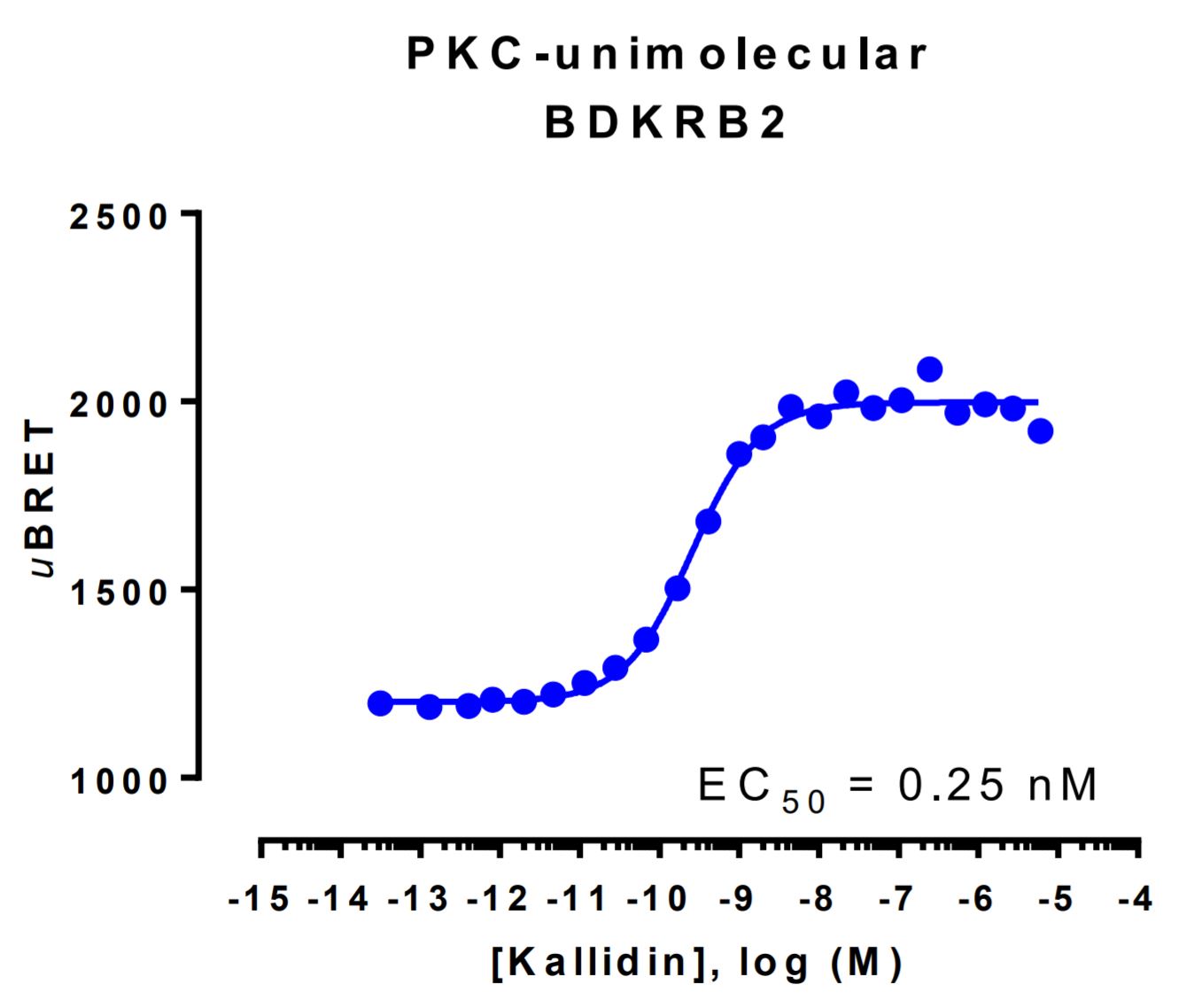
HEK293 cells were transfected with a receptor coding plasmid (the human bradykinin receptor B2 (BDKRB2)) as well as the plasmid coding for the PKC activation biosensor. On the day of BRET, cells were rinsed with assay buffer, incubated with coelenterazine and increasing amounts of kallidin for 10 minutes and BRET subsequently measured.